validity_analysis represents a wrapper function for the bmbstats function.
validity_analysis runs the bootstrap validity analysis of the data data frame using
estimator_function to return the estimators
validity_analysis( data, criterion, practical, SESOI_lower = SESOI_lower_validity_func, SESOI_upper = SESOI_upper_validity_func, estimator_function = validity_estimators, control = model_control(), na.rm = FALSE )
Arguments
| data | Data frame |
|---|---|
| criterion | Character vector indicating column name in the |
| practical | Character vector indicating column name(s) in the |
| SESOI_lower | Function or numeric scalar. Default is |
| SESOI_upper | Function or numeric scalar. Default is |
| estimator_function | Function for providing validity estimators. Default is |
| control | Control object returned from |
| na.rm | Should NAs be removed? Default is |
Value
Object of class bmbstats
Examples
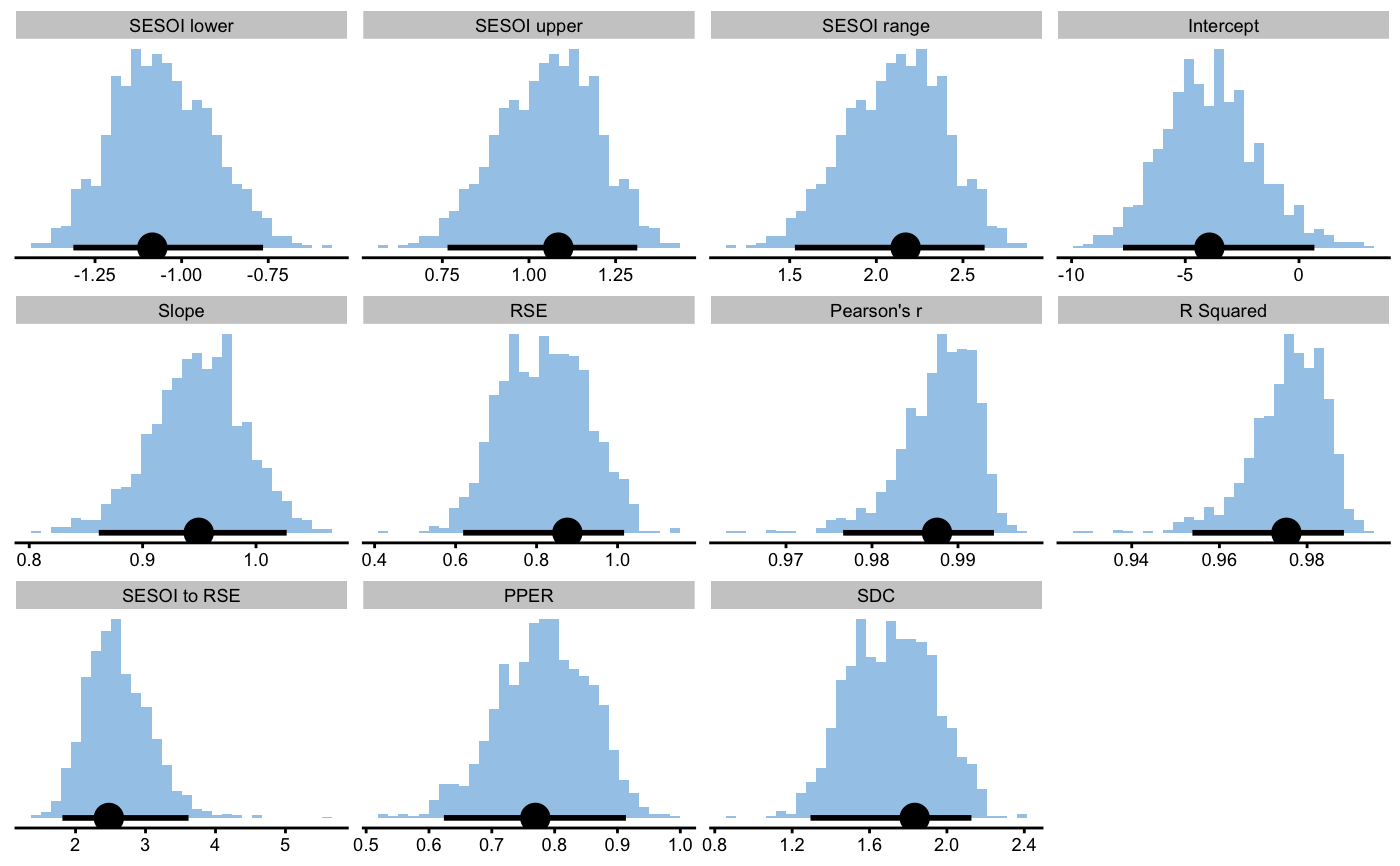
data("agreement_data") val_analysis <- validity_analysis( data = agreement_data, criterion = "Criterion_score.trial1", practical = "Practical_score.trial1", control = model_control( boot_type = "perc", boot_samples = 1000, seed = 1667 ) )#>#>#>val_analysis#> Bootstrap with 1000 resamples and 95% perc confidence intervals. #> #> estimator value lower upper #> SESOI lower -1.0846145 -1.3126039 -0.7649283 #> SESOI upper 1.0846145 0.7649283 1.3126039 #> SESOI range 2.1692290 1.5298567 2.6252079 #> Intercept -3.9394072 -7.7384917 0.6781176 #> Slope 0.9494885 0.8613078 1.0270365 #> RSE 0.8760378 0.6186607 1.0160992 #> Pearson's r 0.9875619 0.9766460 0.9941832 #> R Squared 0.9752786 0.9538374 0.9884002 #> SESOI to RSE 2.4761819 1.8120685 3.6148831 #> PPER 0.7692369 0.6237352 0.9134460 #> SDC 1.8335682 1.2948716 2.1267200plot(val_analysis)