RCT_predict perform additional analysis on model returned by cv_model function
assuming RCT data
RCT_predict( model, new_data, outcome, group, control_label, treatment_label, subject_label = rownames(new_data), na.rm = FALSE )
Arguments
| model | Object returned by |
|---|---|
| new_data | Data frame |
| outcome | Character string indicating the outcome column in |
| group | Character vector indicating the name of the column in |
| control_label | Character vector indicating the label inside the |
| treatment_label | Character vector indicating the label inside the |
| subject_label | Row labels, usually participants names. Default is |
| na.rm | Should NAs be removed? Default is |
Value
Object of class bmbstats_RCT_predict
Examples
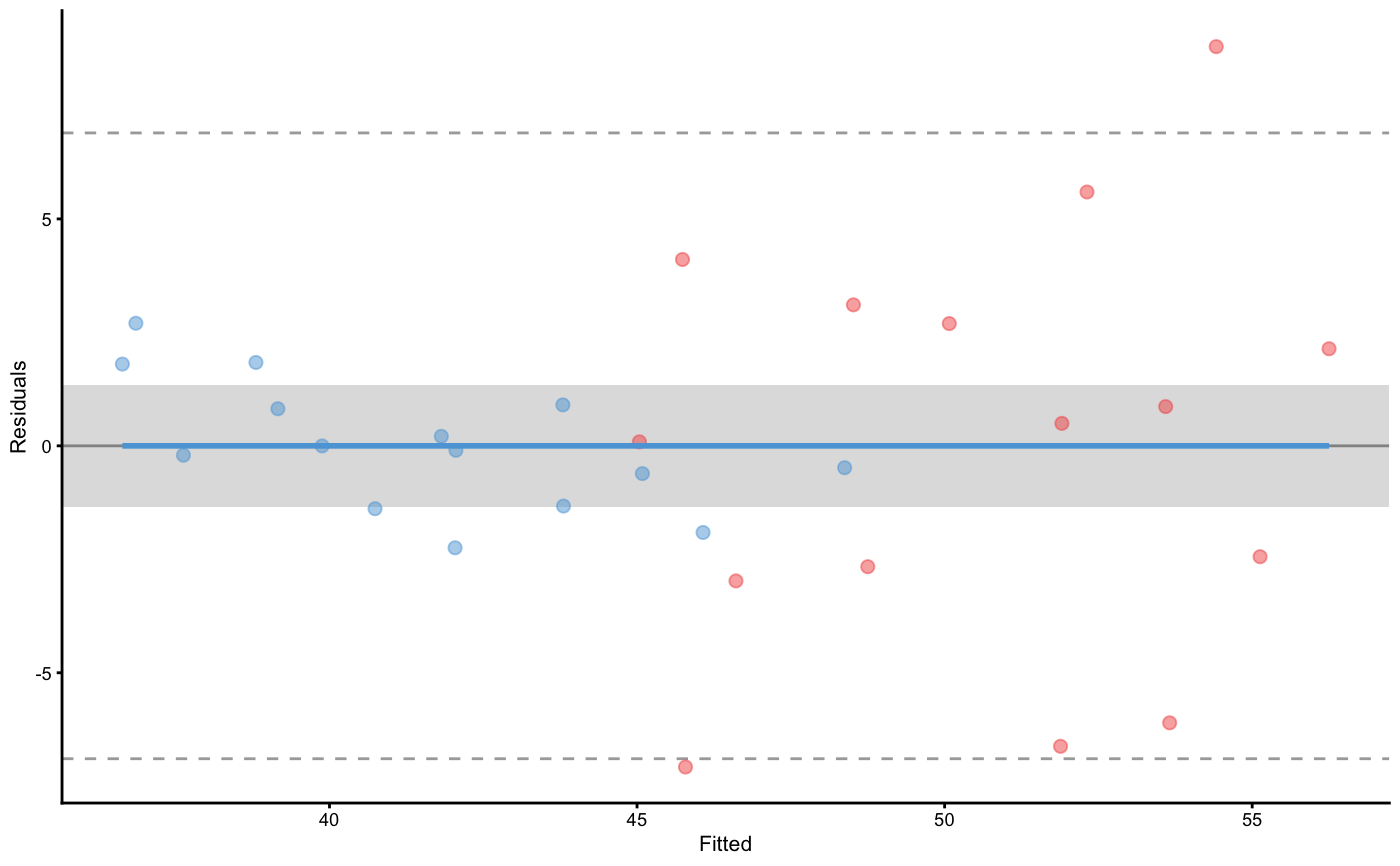
data("vertical_jump_data") m1 <- cv_model( `Post-test` ~ `Pre-test` + Group, vertical_jump_data, control = model_control( cv_repeats = 10, cv_folds = 3, cv_strata = vertical_jump_data$Group ) )#>#>m1_rct <- RCT_predict( m1, new_data = vertical_jump_data, outcome = "Post-test", group = "Group", treatment_label = "Treatment", control_label = "Control" ) m1_rct#> Training data consists of 3 predictors and 30 observations. Cross-Validation of the model was performed using 10 repeats of 3 folds. #> #> Model performance: #> #> metric training training.pooled testing.pooled mean #> MBE -7.342304e-15 -2.723785e-15 0.1033556 0.1033556 #> MAE 2.410129e+00 2.355458e+00 2.8308821 2.8308821 #> RMSE 3.314293e+00 3.202384e+00 3.8552800 3.8101814 #> PPER 3.077021e-01 3.207432e-01 0.2684127 0.2631750 #> SESOI to RMSE 8.130894e-01 8.280495e-01 0.6878184 0.7367233 #> R-squared 7.496390e-01 7.662607e-01 0.6614798 0.6504420 #> MinErr -7.076575e+00 -8.085987e+00 -9.1250789 -6.0198996 #> MaxErr 8.795609e+00 9.666440e+00 11.5199936 6.2312066 #> MaxAbsErr 8.795609e+00 9.666440e+00 11.5199936 8.3183026 #> SD min max #> 1.25564032 -1.77892687 2.4384640 #> 0.49045935 1.64421753 3.5844826 #> 0.59801504 2.17949434 4.8576958 #> 0.06616522 0.19211870 0.4731087 #> 0.19994911 0.53146023 1.3897141 #> 0.17968596 0.04826121 0.8201147 #> 2.37233436 -9.12507894 -1.6820706 #> 3.15085666 1.71461206 11.5199936 #> 1.72624815 4.02865888 11.5199936 #> #> Individual model results: #> #> subject group observed predicted residual magnitude counterfactual #> 1 Control 44.28819 42.04167 -2.246520188 Lower 49.88950 #> 2 Control 47.98205 46.07303 -1.909018621 Lower 53.92086 #> 3 Control 45.13136 43.80419 -1.327164513 Equivalent 51.65202 #> 4 Control 48.85697 48.37440 -0.482576173 Equivalent 56.22222 #> 5 Control 42.12570 40.74050 -1.385202381 Lower 48.58833 #> 6 Control 41.60806 41.81824 0.210179844 Equivalent 49.66606 #> 7 Control 45.69844 45.08679 -0.611653379 Equivalent 52.93462 #> 8 Control 42.89175 43.79421 0.902458750 Equivalent 51.64204 #> 9 Control 42.15402 42.05525 -0.098768933 Equivalent 49.90308 #> 10 Control 37.83235 37.62593 -0.206420330 Equivalent 45.47376 #> 11 Control 34.83170 36.63302 1.801323154 Higher 44.48085 #> 12 Control 39.88302 39.88062 -0.002402407 Equivalent 47.72844 #> 13 Control 38.34326 39.16084 0.817582171 Equivalent 47.00867 #> 14 Control 36.96744 38.80457 1.837133658 Higher 46.65240 #> 15 Control 34.15168 36.85273 2.701049347 Higher 44.70056 #> 16 Treatment 52.86402 45.78745 -7.076574931 Lower 37.93962 #> 17 Treatment 58.50489 51.88463 -6.620263046 Lower 44.03680 #> 18 Treatment 59.76192 53.65842 -6.103493730 Lower 45.81060 #> 19 Treatment 49.58421 46.60804 -2.976168124 Lower 38.76021 #> 20 Treatment 51.41375 48.75083 -2.662918492 Lower 40.90300 #> 21 Treatment 57.57113 55.12834 -2.442796297 Lower 47.28051 #> 22 Treatment 51.41158 51.90657 0.494991154 Equivalent 44.05874 #> 23 Treatment 44.94979 45.03811 0.088313055 Equivalent 37.19028 #> 24 Treatment 52.72907 53.59372 0.864643040 Equivalent 45.74589 #> 25 Treatment 47.38184 50.07579 2.693952355 Higher 42.22797 #> 26 Treatment 54.11023 56.24927 2.139040280 Higher 48.40144 #> 27 Treatment 41.63256 45.73852 4.105951767 Higher 37.89069 #> 28 Treatment 45.41064 48.51731 3.106671448 Higher 40.66948 #> 29 Treatment 46.72143 52.31447 5.593042297 Higher 44.46664 #> 30 Treatment 45.61997 54.41558 8.795609224 Higher 46.56775 #> pITE pITE_magnitude #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> 7.847828 Higher #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> -7.847828 Lower #> #> Summary of residuals per RCT group: #> #> group mean SD #> Control -9.947512e-15 1.426115 #> Treatment -4.736662e-15 4.637293 #> #> Summary of counterfactual effects of RCT group: #> #> group pATE pVTE #> Treatment -7.847828 0 #> Control 7.847828 0 #> pooled 7.847828 0 #> #> Treatment effect summary #> #> Average Treatment effect: 7.847828 #> Variable Treatment effect: 0 #> Random Treatment effect: 4.412559plot(m1_rct)